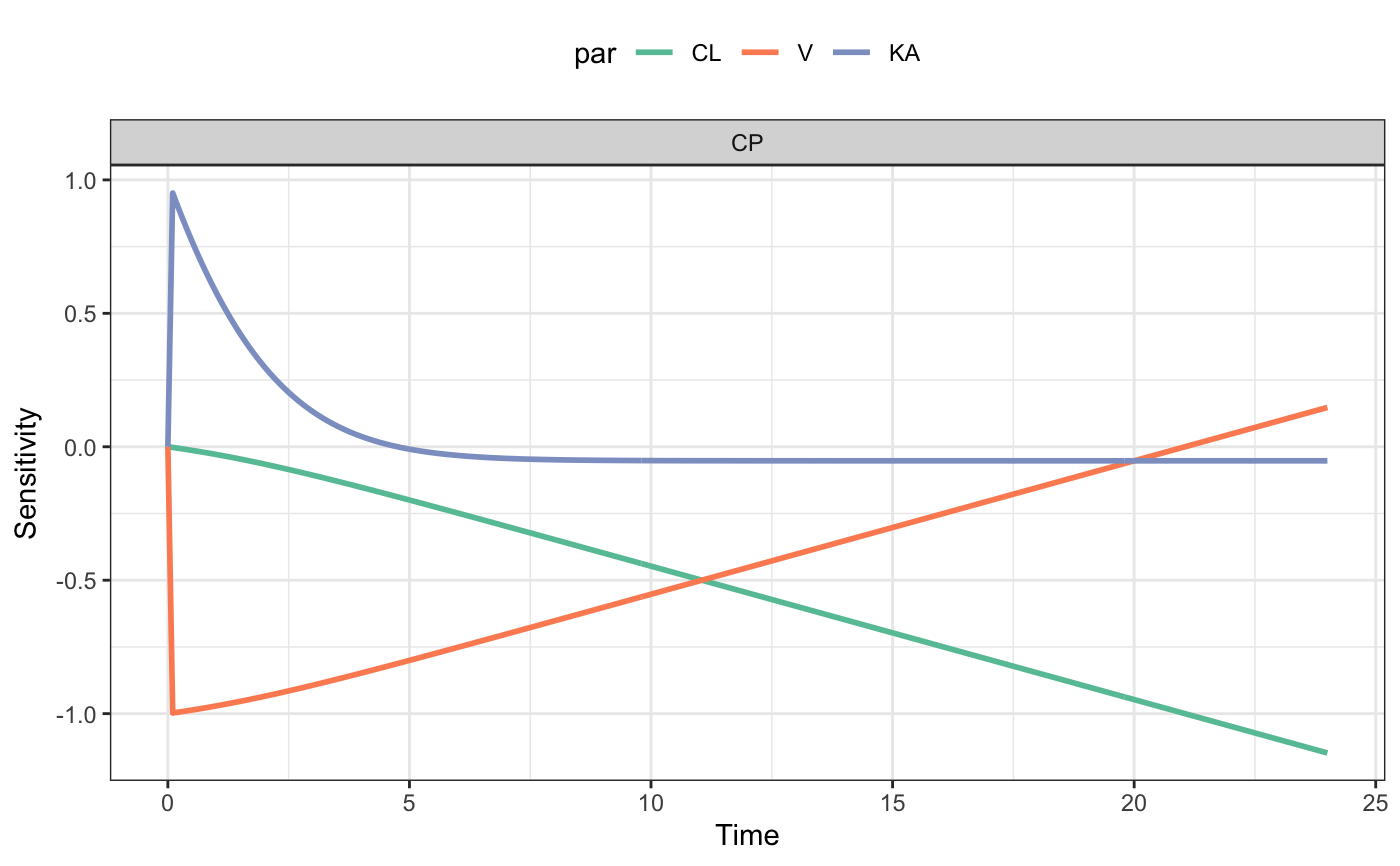
Perform local sensitivity analysis
lsa(mod, fun, par, var, eps = 1e-08, ...)
Arguments
| mod | a mrgsolve model object |
|---|---|
| fun | generating simulated for sensitivity analysis (see details) |
| par | parameter names as character vector or comma-separated string |
| var | output names (compartment or capture) as character vector or comma-separated string |
| eps | parameter change value for sensitivity analysis |
| ... | arguments passed to |
Examples
#>#>par <- "CL,V,KA" var <- "CP" dose <- ev(amt = 100) fun <- function(p) mrgsim_e(mod,dose,param=p,output="df") out <- lsa(mod, fun, par, var) head(out)#> time var value par sens #> 1 0.0 CP 0.0000000 CL 0.000000000 #> 2 0.0 CP 0.0000000 CL 0.000000000 #> 3 0.1 CP 0.4746056 CL -0.002539740 #> 4 0.2 CP 0.9016794 CL -0.005158389 #> 5 0.3 CP 1.2857564 CL -0.007855927 #> 6 0.4 CP 1.6309401 CL -0.010631929plot(out)#>